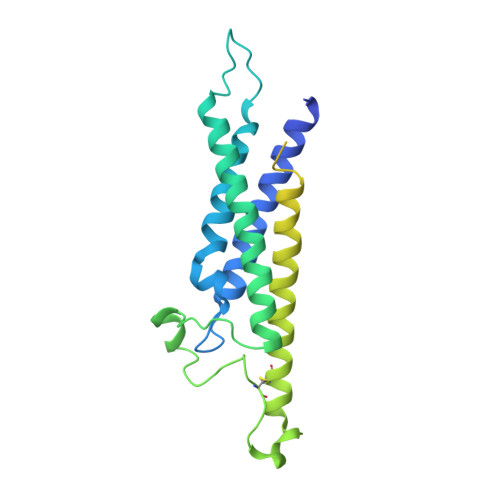
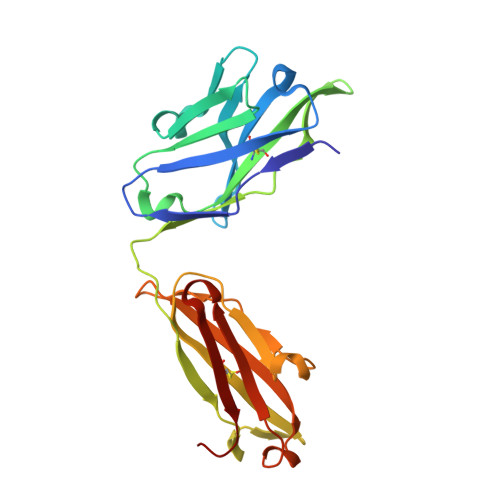
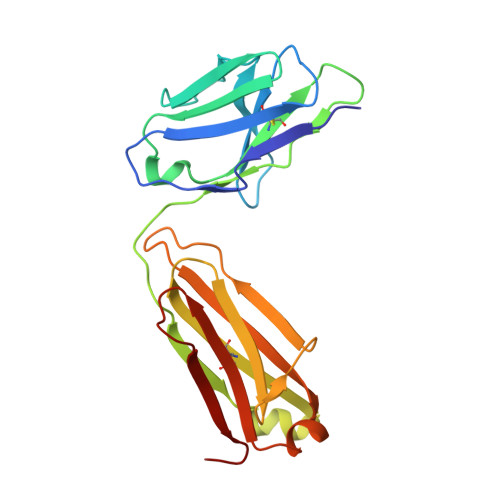
Structure of CD20 in complex with the therapeutic monoclonal antibody rituximab.
Rouge, L., Chiang, N., Steffek, M., Kugel, C., Croll, T.I., Tam, C., Estevez, A., Arthur, C.P., Koth, C.M., Ciferri, C., Kraft, E., Payandeh, J., Nakamura, G., Koerber, J.T., Rohou, A.(2020) Science 367: 1224-1230
- PubMed: 32079680
- DOI: https://doi.org/10.1126/science.aaz9356
- Primary Citation of Related Structures:
6VJA - PubMed Abstract:
Cluster of differentiation 20 (CD20) is a B cell membrane protein that is targeted by monoclonal antibodies for the treatment of malignancies and autoimmune disorders but whose structure and function are unknown. Rituximab (RTX) has been in clinical use for two decades, but how it activates complement to kill B cells remains poorly understood. We obtained a structure of CD20 in complex with RTX, revealing CD20 as a compact double-barrel dimer bound by two RTX antigen-binding fragments (Fabs), each of which engages a composite epitope and an extensive homotypic Fab:Fab interface. Our data suggest that RTX cross-links CD20 into circular assemblies and lead to a structural model for complement recruitment. Our results further highlight the potential relevance of homotypic Fab:Fab interactions in targeting oligomeric cell-surface markers.
- Department of Structural Biology, Genentech Inc., South San Francisco, CA 94080, USA.
Organizational Affiliation: